Japanese / English
Image standardization for generic feature quantification
The applications of whole slide image (WSI) analysis and pattern recognition technologies enable the quantification of features in histopathology, and it is expected to support the diagnosis with the concurrent use of machine learning. Those features will be exploited as morphological or textural biomarkers for what is called ‘morphomics’ or ‘histomics’. However, the results of feature quantification depend on the image quality obtained by WSI scanners, and it is a serious limitation in the application of computerized image analysis [Fig. 1].
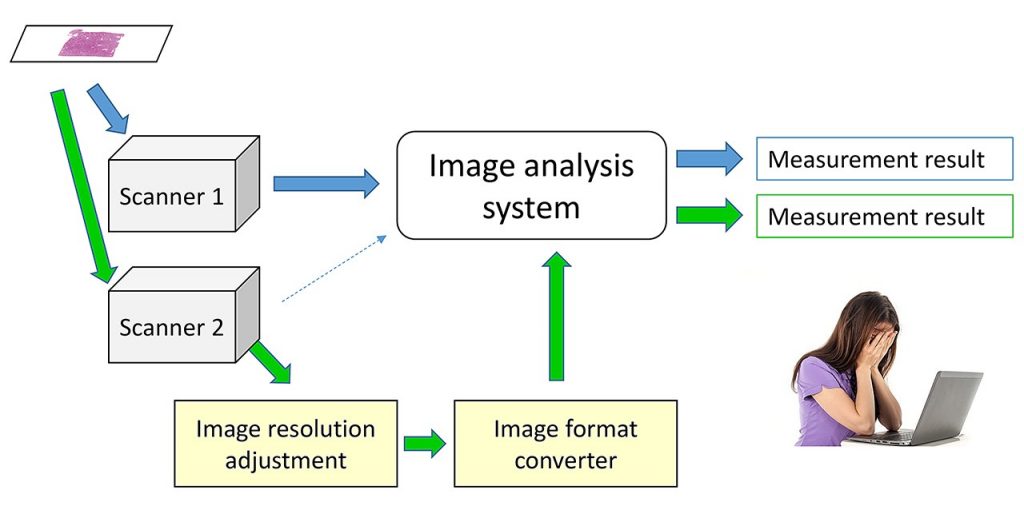
Fig. 1
In this study, we investigate image standardization technology to overcome this limitation. The image standardization consists of the calibration of color and spatial frequency response. The color calibration has been carried out by scanning a color chart slide for device characterization, and/or by color unmixing for correcting the stain variation. The spatial frequency response depends on the modulation transfer function (MTF) and the focusing performance of an imaging device, and is adjusted with digital filtering.
For assessing the effect of image standardization, nuclear and structural features were calculated by automated image analysis with and without standardization. Without image standardization, the results of nuclear segmentation and feature quantification varied on scanners even when the same slide was analyzed, such as nuclear area, shape, and texture. In our preliminary experiment, the variation was decreased and the possibility of feature quantification independent on the imaging device were demonstrated [Fig. 2, Table 1]. However, some features such as texture features are difficult to be standardized.
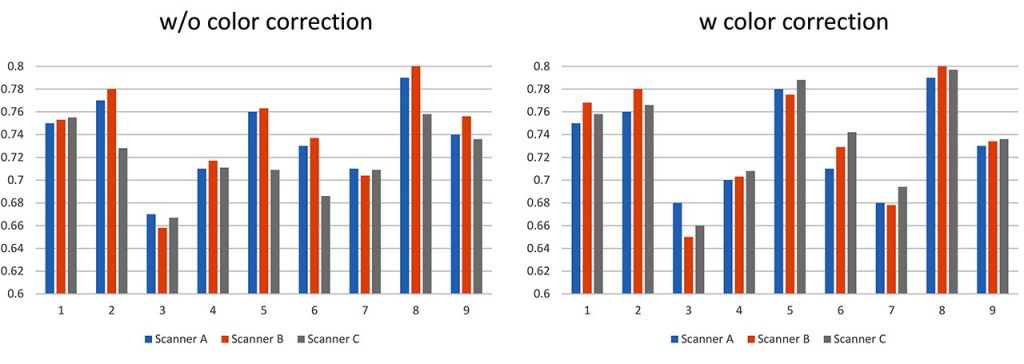
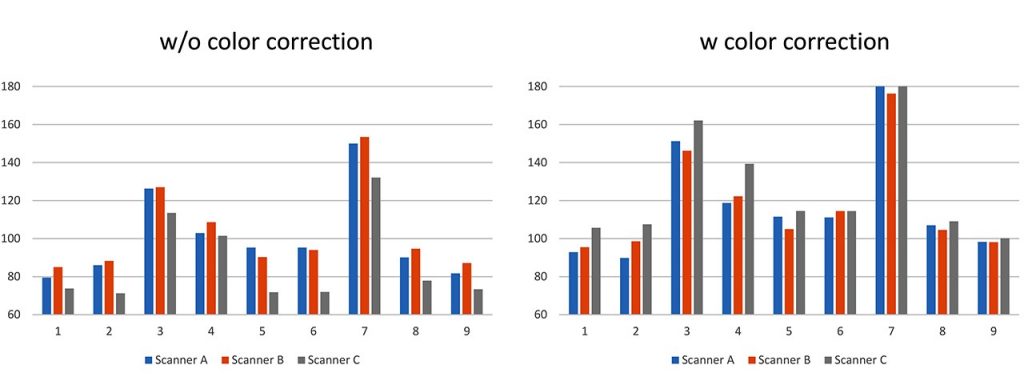
Fig. 2 Feature quantification results from the images acquired by three different scanners, with and without color correction.
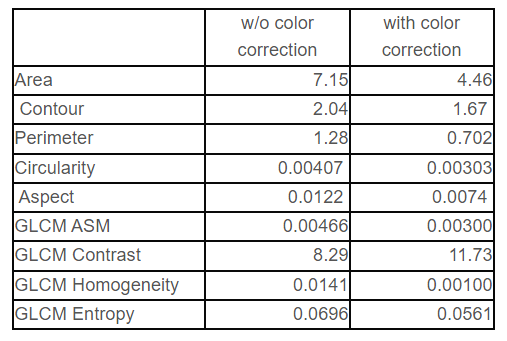
Table 1 Standard deviation among different scanners (average of 9 images).
It is also necessary that the algorism for image analysis and quantification needs to be considered for achieving generic morphological and textural biomarkers usable as the index for diagnosis and prognosis.
This work is partially supported by New Energy and Industrial Technology Development Organization (NEDO), Japan. The author acknowledges the organization and the project members from Keio University, NEC Corporation, Saitama Medical University, and Tokyo Institute of Technology.
Related publications
- Maulana Abdul Aziz, Tomoya Nakamura, Masahiro Yamaguchi, Tomoharu Kiyuna, Yoshiko Yamashita, Tokiya Abe, Akinori Hashiguchi, Michiie Sakamoto. Effectiveness of color correction on the quantitative analysis of histopathological images acquired by different whole-slide scanners, Artificial Life and Robotics, Jul. 2018. [SharedIt ]
- Masahiro Yamaguchi. Toward the generic quantification of morphological and textural features: the impact of image standardization, 3rd Digital Pathology Congress Asia 2017. (2017)
- Yuri Murakami, Tokiya Abe, Akinori Hashiguchi, Masahiro Yamaguchi, Michiie Sakamoto. Color correction for automatic fibrosis quantification in liver biopsy specimens, Journal of Pathology Informatics, 4:36. (2013)
- T. Abe, Y. Murakami, M. Yamaguchi, N. Ohyama, and Y. Yagi, “Color correction of pathological images based on dye amount quantification,” Opt. Rev., 12, 4, 293-300, (2005)
- T. Abe, M. Yamaguchi, Y. Murakami, N. Ohyama,Y. Yagi, “Color correction of pathological images for different staining-condition slides,” IEEE HealthCom 2004, Odawara, Japan, 218-223, (2004)
